EMGM Antibiotic Resistance Working Group
Convener: Muhamed-Kheir Taha, Neisseria Unit, Institut Pasteur, Paris, France, mktaha@pasteur.fr.
Co-convener: Julio Vázquez, Reference Laboratory for Neisserias, National Center for Microbiology, Institute of Health Carlos III, Madrid, Spain, jvazquez@isciii.es.
Since the first EMGM meeting (Rhodes 1991), the working group on antibiotic resistance (Wgatb) started first to explore methods and techniques used by different members of the EMGM.
The aims of the working group on antibiotic resistance are:
- Standardization of methods and break point definitions.
- Implementation and standardization of molecular tools to determine meningococcal susceptibility to antibiotics.
- Implementation of non-culture methods using PCR for detection of antibiotics resistance in clinical samples.
Several contributions have been already achieved:
In 2003: Recommendations on methods and media to be used were made available to the scientific community through the paper:
Vázquez JA , Arreaza L , Block C , Ehrhard I , Gray S, Heuberger S , Hoffmann S , Kriz P , Nicolas P , Olcen P, Skoczynska A , Spanjaard L , Stefanelli P , Taha MK ,Tzanakaki G. Interlaboratory comparison of agar dilution and Etest methods for determining the MICs of antibiotics used in management of Neisseria meningitidis infections. Antimicrob Agents Chemother. 2003 Nov; 47(11):3430-4.
In 2005: Recommendations on the use of PCR-based approaches as a non-culture method to detect isolates with reduced susceptibility to penicillin through the paper:
Taha MK, Zarantonelli ML, Neri A, Enriquez R, Vázquez JA, Stefanelli P. Interlaboratory comparison of PCR-based methods for detection of penicillin G susceptibility in Neisseria meningitidis. Antimicrob Agents Chemother. 2006 Mar; 50(3):887-92.
In 2007: The use of penA sequencing to identify isolates with reduced susceptibility to penicillin G was recommended and a database of penA sequences was established. This data base is accessible by the Internet and can be used in the same way that other similar data bases hosted at neisseria.org. Several examples of the access to the penA data base:
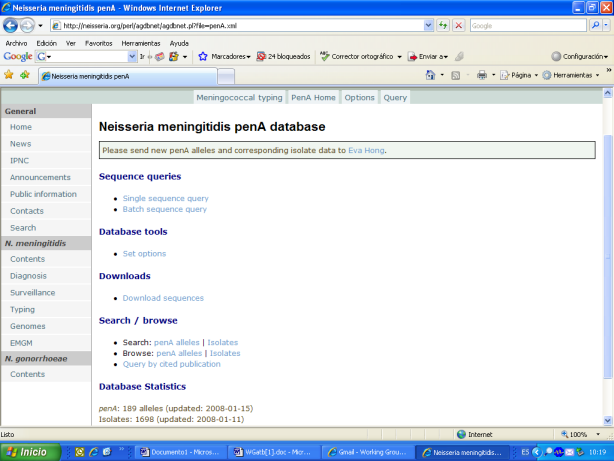
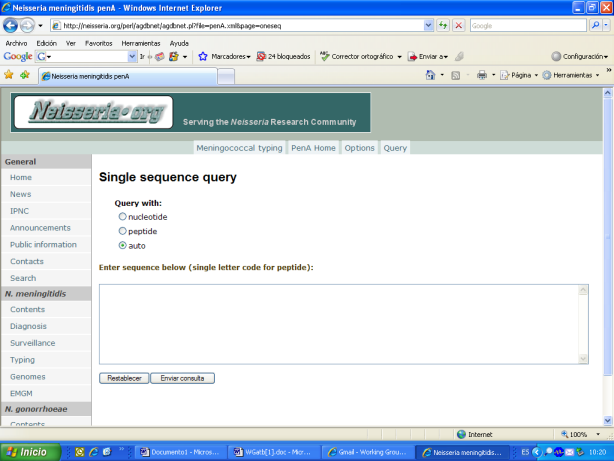
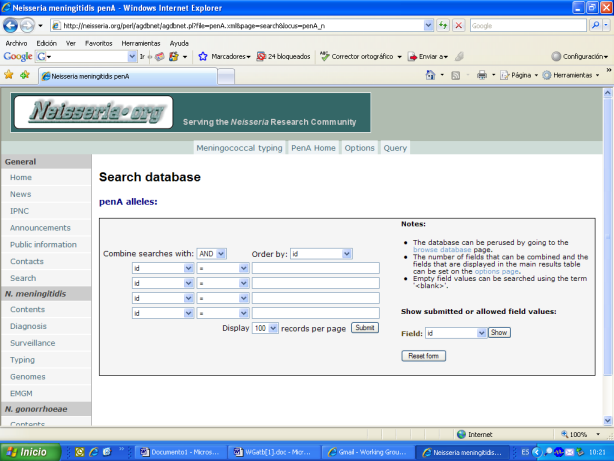
Data were published:
Taha MK, Vázquez JA, Hong E, Bennett DE, Bertrand S, Bukovski S, Cafferkey MT, Carion F, Christensen JJ, Diggle M, Edwards G, Enríquez R, Fazio C, Frosch M, Heuberger S, Hoffmann S, Jolley KA, Kadlubowski M, Kechrid A, Kesanopoulos K, Kriz P, Lambertsen L, Levenet I, Musilek M, Paragi M, Saguer A, Skoczynska A, Stefanelli P, Thulin S, Tzanakaki G, Unemo M, Vogel U, Zarantonelli ML. Target gene sequencing to characterize the penicillin G susceptibility of Neisseria meningitidis. Antimicrob Agents Chemother. 2007 51(8):2784-92.
Currently, the WG carry out a new collaborative study on the molecular characterization of meningococcal resistance to rifampicin and ciprofloxacin through the sequencing of rpoB, gyrA, parE and parC.
